mygrad.nnet.layers.conv_nd#
- mygrad.nnet.layers.conv_nd(x: ArrayLike, filter_bank: ArrayLike, *, stride: int | Tuple[int, ...], padding: int | Tuple[int, ...] = 0, dilation: int | Tuple[int, ...] = 1, constant: bool | None = None) Tensor[source]#
Use
filter_bank(w) to perform strided N-dimensional neural network-style convolutions (see Notes) overx.:f(x, w) -> x ⋆ w shapes: (N, C, X0, ...) ⋆ (F, C, W0, ...) -> (N, F, G0, ...)
xrepresents a batch of data over which the filters are convolved. Specifically, it must be a tensor of shape \((N, C, X_0, ...)\), where \(N\) is the number of samples in the batch, C is the channel-depth of each datum, and \((X_0, ...)\) are the dimensions over which the filters are convolved. Accordingly, each filter must have a channel depth of \(C\).Thus convolving \(F\) filters, each with a shape \((C, W_0, ...)\), over the data batch will produce a tensor of shape \((N, F, G_0, ...)\), where \((G_0, ...)\) is the shape of the grid commensurate with the filter placements
- Parameters:
- xArrayLike, shape=(N, C, Xo, …)
The data batch to be convolved over.
- filter_bankUnion[Tensor, array_like], shape=(F, C, Wo, …)
The filters used to perform the convolutions.
- strideUnion[int, Tuple[int, …]]
(keyword-only argument) The step-size with which each filter is placed along the H and W axes during the convolution. The tuple indicates (stride-0, …). If a single integer is provided, this stride is used for all convolved dimensions
- paddingUnion[int, Tuple[int, …]]
(keyword-only argument) The number of zeros to be padded to both ends of each convolved dimension, respectively. If a single integer is provided, this padding is used for all of the convolved axes
- dilationUnion[int, Tuple[int, …]], optional (default=1)
(keyword-only argument) The spacing used when placing kernel elements along the data. E.g. for a 1D convolution the ith placement of the kernel multiplied against the dilated-window:
x[:, :, i*s:(i*s + w*d):d], wheresis the stride,wis the kernel-size, anddis the dilation factor.If a single integer is provided, that dilation value is used for all of the convolved axes
- constantOptional[None]
If True, the resulting Tensor is a constant.
- Returns:
- Tensor, shape=(N, F, G0, …)
The result of each filter being convolved over each datum in the batch.
Notes
The filters are not flipped by this operation, meaning that an auto-correlation is being performed rather than a true convolution.
Only ‘valid’ filter placements – where the filters overlap completely with the (padded) data – are permitted.
Examples
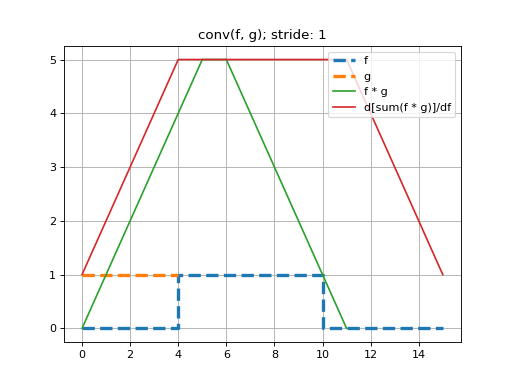
Here we perform a 1D convolution of a constant-valued kernel,
k, with a ‘square-wave’ signal,x, using stride-1. Note that because we are constrained to doing deep learning-style convolutions, that we prepend the dimensions \((N=1, C=1)\) tox, and \((F=1, C=1)\) and tok. That is, we are performing a convolution on one, single-channeled signal using one kernel.See that this convolution produces the expected triangle-shaped response. The shape of the resulting tensor is \((N=1, F=1, G_0=12)\). That is, the length-5 kernel can be placed in 12 valid positions, using a stride of 1.
>>> import mygrad as mg >>> from mygrad.nnet import conv_nd >>> x = mg.zeros((1, 1, 16)) # a square-wave signal >>> x[..., 5:11] = 1 >>> k = mg.ones((1, 1, 5)) # a constant-valued kernel >>> conv_nd(x, k, stride=1) # performing a stride-1, 1D convolution Tensor([[[0., 1., 2., 3., 4., 5., 5., 4., 3., 2., 1., 0.]]], dtype=float32)
Back-propagating through the (summed) convolution:
>>> conv_nd(x, k, stride=1).sum().backward() # sum to a scalar to perform back-prop >>> x.grad # d(summed_conv)/dx array([[[1., 2., 3., 4., 5., 5., 5., 5., 5., 5., 5., 5., 4., 3., 2., 1.]]], dtype=float32) >>> k.grad # d(summed_conv)/dk array([[[6., 6., 6., 6., 6.]]])
(Source code, png, hires.png, pdf)
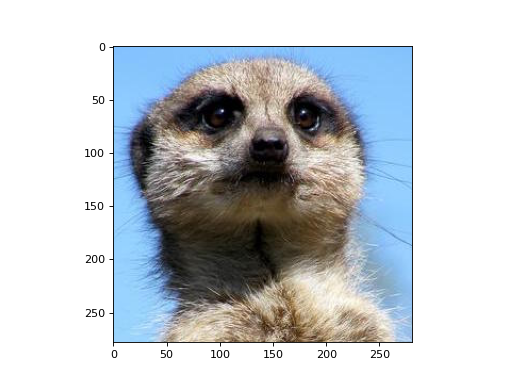
Let’s apply a edge-detection kernel to each color channel of an RGB image.
>>> import matplotlib.pyplot as plt >>> import matplotlib.image as mpimg >>> from mygrad.nnet.layers import conv_nd >>> # A shape-(H, W, 3) RGB image >>> img = mpimg.imread('../_static/meerkat.png') >>> # We'll treat this like a batch of three greyscale images >>> # where each "image" is actually a color channel >>> # shape-(H, W, 3) -> shape-(3, 1, H, W) >>> x = img.transpose(2, 0, 1)[:, None, :, :]
>>> # edge detection kernel >>> kernel = np.array([[-1, -1, -1], ... [-1, 8, -1], ... [-1, -1, -1]]) >>> # (Hf, Wf) --> (1, 1, Hf, Wf) >>> kernel = kernel.reshape(1, 1, *kernel.shape)
>>> # conv: (3, 1, H, W) w/ (1, 1, Hf, Wf) --> (3, 1, H', W') >>> # squeeze + transpose: (3, 1, H', W') --> (H', W', 3) >>> processed = conv_nd(x, kernel, stride=(1, 1)) >>> processed = processed.data.squeeze().transpose(1, 2, 0)
>>> fig, ax = plt.subplots() >>> ax.imshow(img)
>>> fig, ax = plt.subplots() >>> ax.imshow(processed)
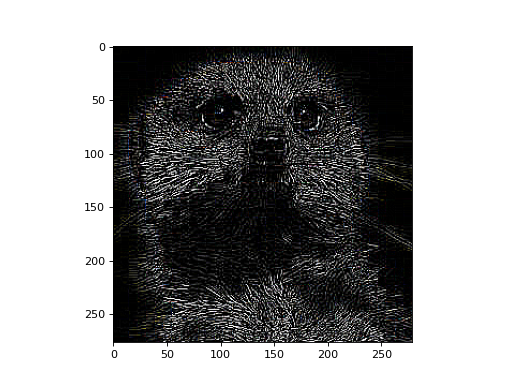
Now, let’s demonstrate a more typical usage for
conv_ndin the context of neural networks.xwill represent 10, 32x32 RGB images, and we will use 5 distinct 2x2 kernels to convolve over each of these images . Note that each kernel must possess 3-channel - one for each RGB channel.That is, we will be performing NxF channel-wise 2D convolutions. Supposing that we don’t want the kernel placements to overlap, we can use a stride of 2. In total, this will produce a shape-\((N=10, F=5, G_0=16, G_1=16)\) tensor as a result.
>>> import mygrad as mg >>> x = mg.random.rand(10, 3, 32, 32)) # creating 10 random 32x32 RGB images >>> k = mg.random.rand(5, 3, 2, 2)) # creating 5 random 3-channel 2x2 kernels
Given the shapes of
xandk,conv_ndautomatically executes a 2D convolution:>>> conv_nd(x, k, stride=2).shape (10, 5, 16, 16)
Extrapolating further,
conv_ndis capable of performing ND convolutions!Performing a convolution over a batch of single-channel, “spatial-3D” tensor data:
>>> # shape-(N=1, C=1, X=10, Y=12, Z=10) >>> x = mg.random.rand(1, 1, 10, 12, 10) >>> # shape-(F=2, C=1, Wx=3, Wy=1, Wz=2) >>> k = mg.random.rand(2, 1, 3, 1, 32) >>> conv_nd(x, k, stride=1).shape (1, 2, 8, 12, 9)